
Congratulations to Dr. Xiaolu Wei for successfully defending her thesis! Dr. Wei gave an awesome talk on meiotic drive and satellite DNA regulation in Drosophila melanogaster. We are so proud of her!

What a whirlwind the past couple of years has been! Check out what we’ve been up to in 2021 and 2022:




Congratulations to team supergene–Sherif Negm (team captain), Xiaolu Wei, John Sproul and Lucas Hemmer— for coming in 2nd place in the open division of the GIDS biomedical data science hackathon! Way to represent the lab and the Bio Department.

 Xiaolu Wei is a recipient of an Agnes M. Messersmith and George Messersmith Dissertation Fellowship – a highly competitive fellowship that recognizes promising University of Rochester PhD candidates. This fellowship will support Xiaolu’s dissertation work on satellite DNA regulation and Segregation Distorter in 2020-2021. Congratulations Xiaolu!
Xiaolu Wei is a recipient of an Agnes M. Messersmith and George Messersmith Dissertation Fellowship – a highly competitive fellowship that recognizes promising University of Rochester PhD candidates. This fellowship will support Xiaolu’s dissertation work on satellite DNA regulation and Segregation Distorter in 2020-2021. Congratulations Xiaolu!

Ching-Ho received a DeLill Nasser award for professional development in genetics to travel to TAGC this spring. This award is in memory of DeLill Nasser, a long time advocate for early career geneticists.
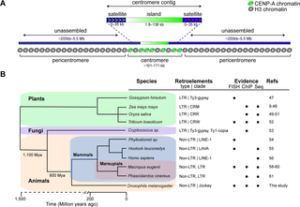
Check out our new paper on the organization of Drosophila melanogaster centromeres. For this paper, our lab collaborated with Barbara Mellone’s lab (University of Connecticut) and Ting Wu’s lab (Harvard University) to discover all of the centromeres in D. melanogaster. Centromeres are genomic regions essential for proper chromosome segregation during cell division. They also are among the most rapidly evolving regions of genomes and can play a role in karyotype evolution and speciation. We know little about the detailed organization of centromeres because they are buried in large blocks of tandem repeats that are difficult to assemble. We combined Ching-Ho’s heterochromatin-enriched assembly methods, with ChIP-seq for the centromere-specific histone variant, CENP-A, and FISH on stretched chromatin fibers to reveal the DNA sequences underlying centromeres. It turns out that centromeres correspond to islands rich in retroelements embedded in a sea of satellite DNA! One retroelement, G2/Jockey-3, is found at all D. melanogaster centromeres and centromeres in D. simulans.
Retroelements are associated with centromeres in a wide range of organisms including fungi, plants, and mammals. This suggests that retroelements may be conserved features of centromeres and may have a role in centromere function. We’re currently working on studying patterns of polymorphism and divergence in G2/Jockey-3 and centromere islands to see how they contribute to centromere evolution and function.
Taylar successfully defended her senior thesis on suppressors of Segregation Distorter in Drosophila melanogaster and is graduating with honors in research this weekend! Next up for Taylar is the Cell, Development, Molecular Biology and Biophysics PhD program Johns Hopkins, where she will be a Morgan Fellow. Congratulations all around Taylar!