All posts by Amanda Larracuente
Ching-Ho wins a scholarship
Firefly chromosomes
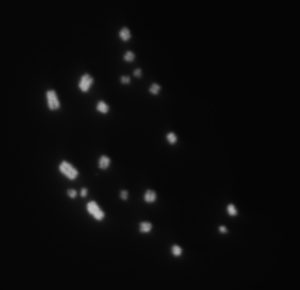
Firefly season is over. It only lasts a couple of months here in Rochester. We rear fireflies in the lab over the summer to study the chromosomal distributions of their repeats. This is challenging for a number of reasons, one being that Photinus pyralis chromosomes typically appear as amorphous little blobs under the microscope. We spend a few months of each summer trying to find good ways to image chromosomes in the short time that we have with the fireflies. In the last week of this summer, Isaac Wong (an undergrad in the lab) optimized protocols to find chromosomes – and they don’t look like amorphous blobs!
A few weeks later, UR wrote an article on fireflies that mentions our work. You can read it here: http://www.rochester.edu/newscenter/firefly-researchers-mapping-worlds-second-most-interesting-genome-269372/.
Congratulations graduates!
Two fantastic undergrads in the lab graduated and are moving on to bigger and better things. Kwan graduated with BS degrees in Computational Biology and Data Science. Alex graduated with a BS in Neuroscience with minors in Classical Civilization, Computational Biology, and Psychology as a Social Science. We wish them the best!
Congratulations Ching-Ho
Postdoc in evolutionary genomics:
Christian graduated
This week marks a big milestone for the Larracuente Lab. Our first undergraduate, Christian Silva, has left the nest. Christian pioneered firefly work in the lab and will be sorely missed. He’s off to UC Davis, where he will start his graduate career in plant biology this fall. Good luck, Christian!
Firefly genome project is a SMRT grant finalist!
We are fortunate to be part of a diverse group of biologists collaborating to sequence the genome of Photinus pyralis- the Big Dipper Firefly. Our firefly genome project was chosen as finalist for PacBio’s “Explore your most interesting genome contest”. The winner will be decided by popular vote! If you’d like to support our efforts to sequence the firefly genome, please visit our page on Experiment.com and vote. See details below: 
Fireflies! These silent fireworks on warm summer nights fill us with wonder. But so much about these fascinating critters remains shrouded in mystery- from the details of how they light up their lanterns to the way some species are disappearing. A team of biologists has joined forces to sequence the genome of the Big Dipper Firefly, Photinus pyralis, your common backyard firefly. This project is part of the PacBio “Explore your most interesting genome” contest. If you’d like to see the firefly genome sequenced, please go to http://www.pacb.com/smrtgrant/ and vote for the project. Voting opens Monday, April 11 and runs through Sunday, May 1st. One person can vote once per day. Help us shed light on the genome of these captivating creatures!
Review on the ecology and evolutionary dynamics of meiotic drive
This summer, over 20 biologists studying meiotic drive in a variety of taxa (including flies, plants, mice, birds and fungi) gathered in the Swiss Alps for a workshop to discuss the outstanding questions on meiotic drive. We were thrilled to be a part of this group. One product of this workshop was a review on the ecology and evolutionary dynamics of drive. You can read the review here:
http://www.cell.com/trends/ecology-evolution/abstract/S0169-5347%2816%2900043-4
“Mysteries of the Genome”
Amanda recently presented at Rochester’s Science Café at the Pittsford Barnes and Nobles on the “Mysteries of the genome”. Click here to learn more about the series.
Mysteries of the genome 11/24/15 Less than 2% of the DNA in humangenomes contains conventional genes that code for proteins. Scientists have known for decades that non-protein-coding regions in the genome can have important functions in regulating genes. However, much of the remaining non-coding and non-regulatory DNA consists of sequences frequently referred to as “junk”. This category includes repeated sequences that we know little about but have links to cancer and other diseases. These repetitive genomic regions can go awry causing catastrophic genomic rearrangements and some can even behave like viruses. In this science cafe, we’ll explore the mysterious parts of genomes rich in so-called “junk” DNA. We’ll discuss the controversy over what is considered a “functional genetic element” and how studying genomic differences within and between species helps shed light on a sequence’s functional significance.