
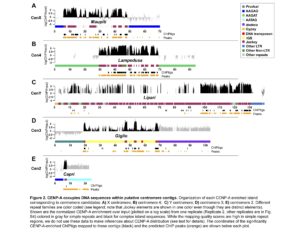
We are interested in the evolution of centromere organization in Drosophila. Centromeres vary in size and complexity across organisms ranging from a single nucleosome in budding yeast to large alpha satellite enriched centromeres in human (Allshire and Karpen 2008). We currently know little about the detailed organization of most eukaryotic centromeres because they are typically buried in repeat-dense heterochromatic regions and are rapidly evolving. We recently collaborated with Barbara Mellone’s lab at the University of Connecticut to combine single molecule long read sequencing, ChIPseq with the centromeric histone variant (CENP-A), and extended chromatin fiber fluorescence in situ hybridization (FISH) to reveal the detailed organization of centromeres in Drosophila melanogaster. We discovered that centromeres are islands of transposable element (TE)-rich DNA embedded within large blocks of tandemly repeated satellite DNAs. Our current projects involve studying how these centromeres evolve over short evolutionary timescales, and particularly, how transposable elements shape molecular and evolutionary aspects of centromere organization.
Representative publications:
Lucas W Hemmer, Sherif Negm, Xuewen Geng, Cécile Courret, Beatriz Navarro-Domínguez, Iain Speece, Xiaolu Wei, Eddyson Altidor, James Chaffer, John S. Sproul, Amanda M Larracuente. 2022. Centromere-associated retroelement evolution in Drosophila melanogaster reveals an underlying conflict. BioRxiv https://doi.org/10.1101/2022.11.25.518008.
Chang, C-H, Chavan, A, Palladino, J., Wei, X., Martins, N.M.C., Santinello, B., Chen, C-C, Erceg, J., Beliveau, B.J., Wu, C-T, Larracuente, A.M., and Mellone, B.G. 2019. Islands of retroelements are major components of Drosophila melanogaster centromeres. BioRxiv Epub 2/1/2019. PLoS Biology https://doi.org/10.1371/journal.pbio.3000241. Recommended by F1000Prime.
This work is supported by an NSF CAREER (MCB) award to AML.
Collaborators: Mellone Lab, Wu Lab
