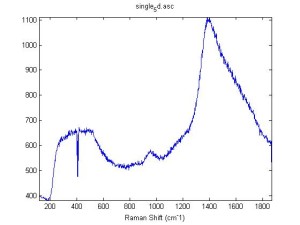
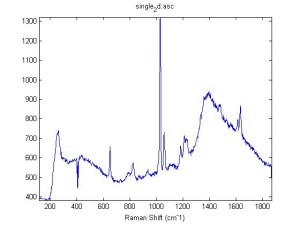
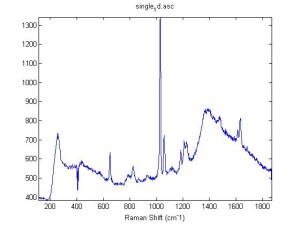
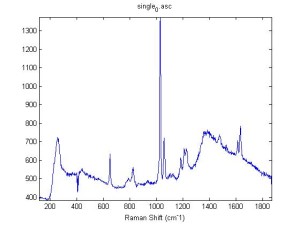
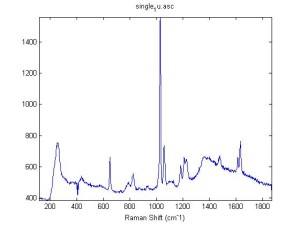
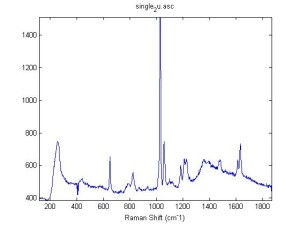
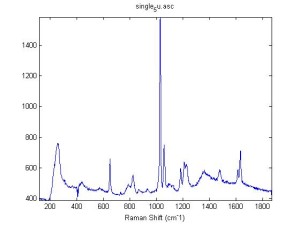
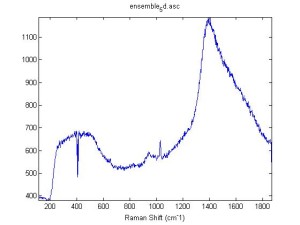
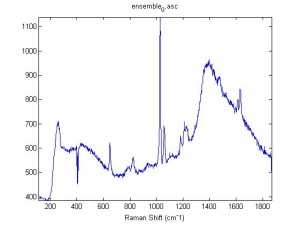
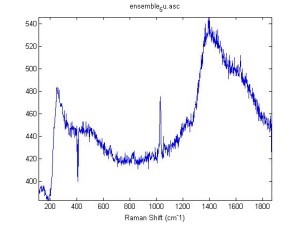
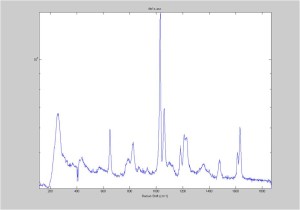
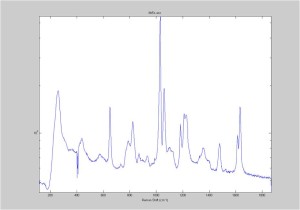
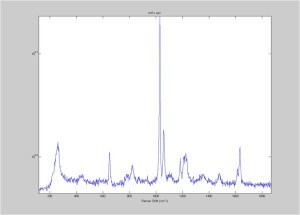
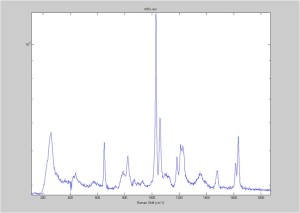
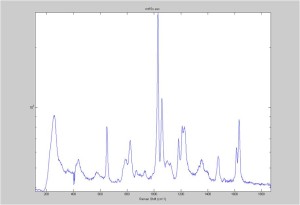
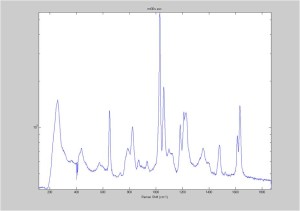
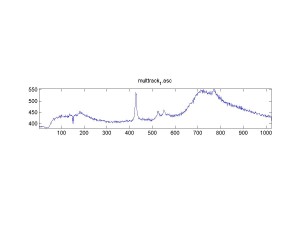
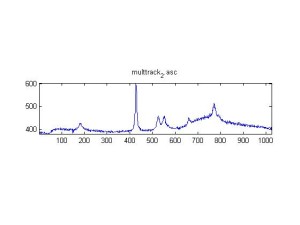
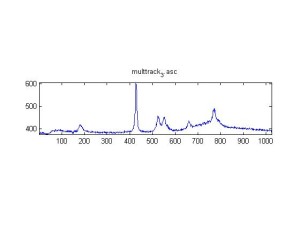
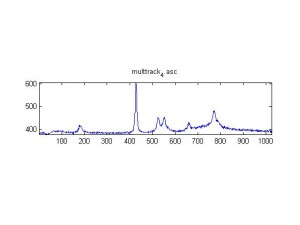
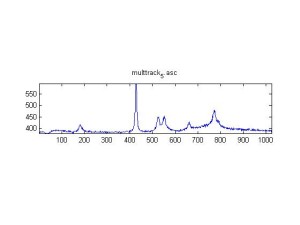
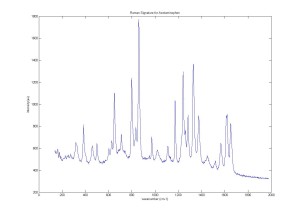
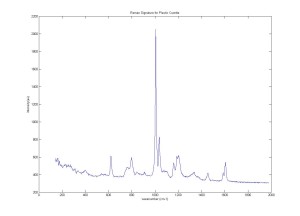
Raman Spectra of 1um beads taken at 20s exposure times. Spectra were taken at different axial positions of trapped bead(s?) and a group of beads that were adhered to the cover slip. These positions ranged from about 5 (microns?) above and below the cover slip (full range is shown for individual beads while select points are shown for the ensemble). The “glass edge” on the right is dominant in all of the adhered ensemble spectra while it disappears as the focus moved farther into the sample for the trapped bead.
Some sources of uncertainty are: 1)finding a place where a bead could be trapped and wouldn’t get knocked out by other beads coming into the trap or leave on its own (I had to start 5 microns above the coverslip to get a stable trap). 2)I used the back reflection of the glass on the microscope’s ccd as a reference point. When I was finishing measurements, I noticed the position of the back reflection was not where I originally set my “0” point (This is not the first time this has been noticed, but I wanted to make a note of it here) 3)I’m pretty sure that 1 increment on the fine tuning knob (which is what I was using to move the objective) corresponds to 1 micron, but I haven’t been able to 100% verify this. 4)Looking at published spectra, the pixel to wavenumber calibration may be slightly off (peaks are shifted to the right a bit). 5)This spectra was taken after looking for and reducing sources of power loss in the Raman path. Part of the process involved adjusting a filter that cleans up the laser light and that may explain the floor around 200 cm^-1