The goal of this project is to use angular scattering from single cells to extract information about sub-cellular scatterers. This could be useful to biologists interested in studying how individual cells respond by detecting changes or differences in the size and shape of organelles.
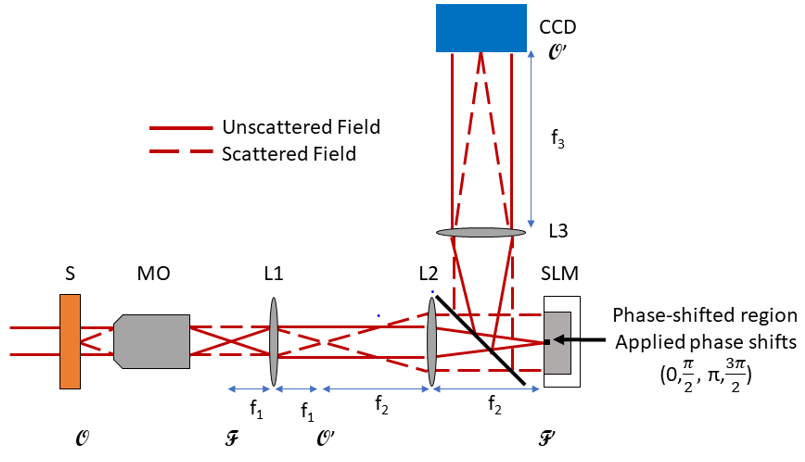
Our imaging system employs Fourier phase microscopy (FPM), which uses the unscattered light as a reference wave for phase shifting interferometry (PSI). Four different phase shifts are applied where the unscattered light hits the center of the spatial light modulator (SLM).
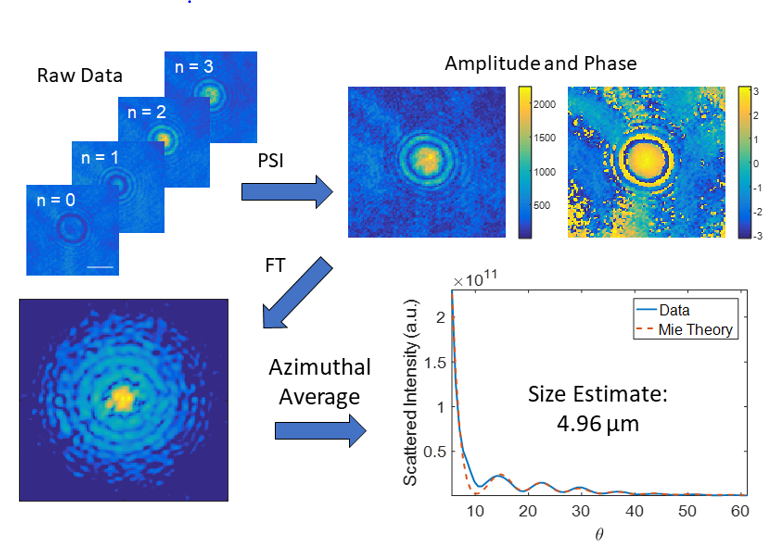
These four PSI images are combined to create the complex field (amplitude and phase) of one quantitative phase image (QPI). This is then Fourier transformed to the angular scattering / Fourier domain. An azimuthal average here tells us how much light scatters at each angle. Mie theory can then be fit to the data to provide a size estimate.
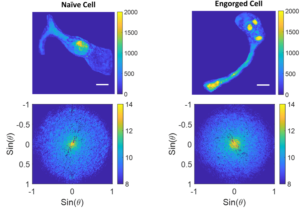
Below are some examples of angular scattering analysis on two different cells, one that has not been exposed to thymocytes, and one that has engorged some thymocytes. Note that the visible thymocytes in the engorged cell correspond to more low angle scattering (the bright spot at 0 degrees).
Future directions of this work include using different angles of illumination to reduce speckle in the angular domain, as well as to provide 3D information about the sample.