Speciation genetics
We study the molecular and evolutionary genetics of the origin of new species, focusing on the evolution of interspecific genetic incompatibilities that cause sterility or inviability in species hybrids. Our long-term goal is to identify and characterize a large panel of hybrid incompatibility genes, providing the material to allow broad generalizations about the evolution of postzygotic reproductive isolation. One of the surprises from our work (and others’) is that speciation and gene flow in Drosophila are mediated by evolutionary arms races between selfish genetic elements— like cryptic drive systems and retrotransposons— and host genomes (Presgraves 2010; Meiklejohn et al. 2018).
Sex chromosomes and speciation
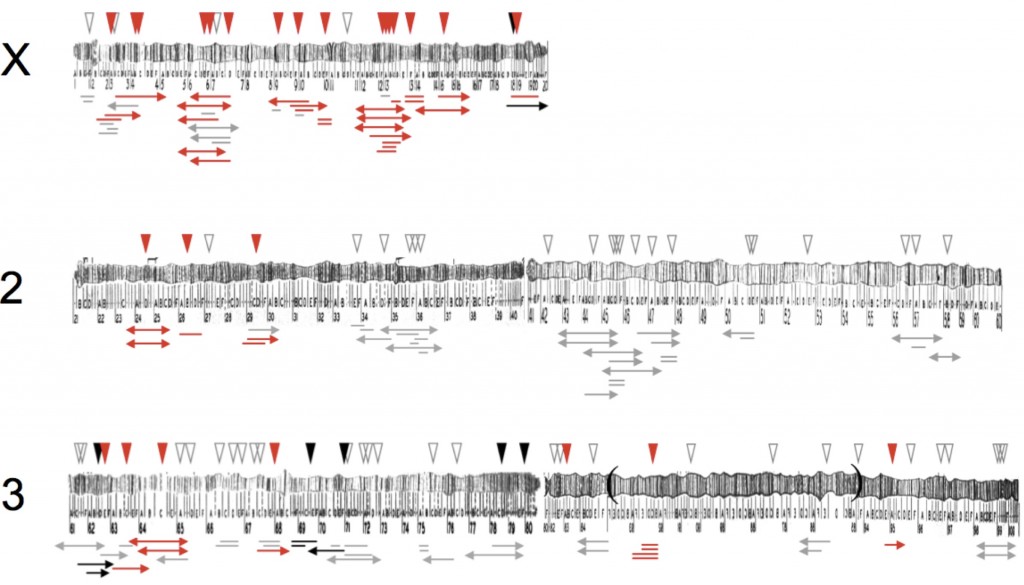
Hybrid sterility is characterized by two strong patterns: Haldane’s rule— the preferential sterility of XY (or ZW) hybrids— and the large X-effect— the disproportionately large effect of the X chromosome in classical genetic analyses of hybrid sterility. In genome-wide fine-scale genetic analyses, we have found that hybrid male sterility genes can accumulate between species ~4x faster on the X chromosome than the autosomes. A major goal of our work now is to determine why the X is a hotspot for the evolution of hybrid sterility. To that end, we are (1) working to identify several X-linked hybrid male sterility factors and (2) studying the regulation of the X chromosome in the Drosophila male germline and its possible disruption in hybrids.
Regulation of the X chromosome in the Drosophila germline
We set out to test the hypotheses that male germline sex chromosome dosage compensation and/or meiotic sex chromosome inactivation (MSCI) might be disrupted in species hybrids, possibly contributing to the high density of hybrid male sterility factors on the X. To our surprise, we discovered that there is little evidence for either process in wildtype pure species flies (Meiklejohn et al. 2012). Instead, an altogether uncharacterized type of transcriptional suppression occurs on the X chromosome. We are working to characterize this process and its genetic control (Landeen et al. 2016).
Genomics of complex speciaton
We use population genomics to study the speciation history of the Drosophila simulans clade species— the cosmopolitan species D. simulans and its two island endemic sister species, D. mauritiana and D. sechellia. Despite significant geographic and reproductive isolation, our analyses reveal that speciation was complex, involving past natural hybridization and introgression. The signal of gene flow is much reduced on the X chromosome, consistent with the notion that interspecific incompatibilities limit genetic exchange between species. Our work, in collaboration with the Meiklejohn lab, integrates genetic mapping and population genomic data to study if and how individual hybrid incompatibility genes affect local gene flow.We study how coevolutionary arms between host genomes and their selfish genes contribute speciation.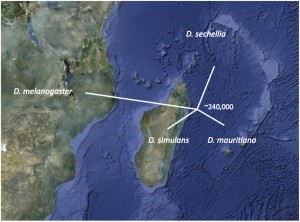
The selfish Segregation Distorter complex
The ultraselfish SD gene complex in Drosophila melanogaster is one of the best-characterized meiotic drive systems. But many mysteries remain about its evolutionary history, its population dynamics, and the functional interactions among the driver (Segregation distorter), target (Responder), and many genetic modifiers. We are now using genetics, molecular biology, population genomics and cytology to investigate SD in populations from France and Africa (Presgraves et al. 2009). See our recent review for more details (Larracuente and Presgraves 2012).
The genetics and evolution of recombination rate differences between species
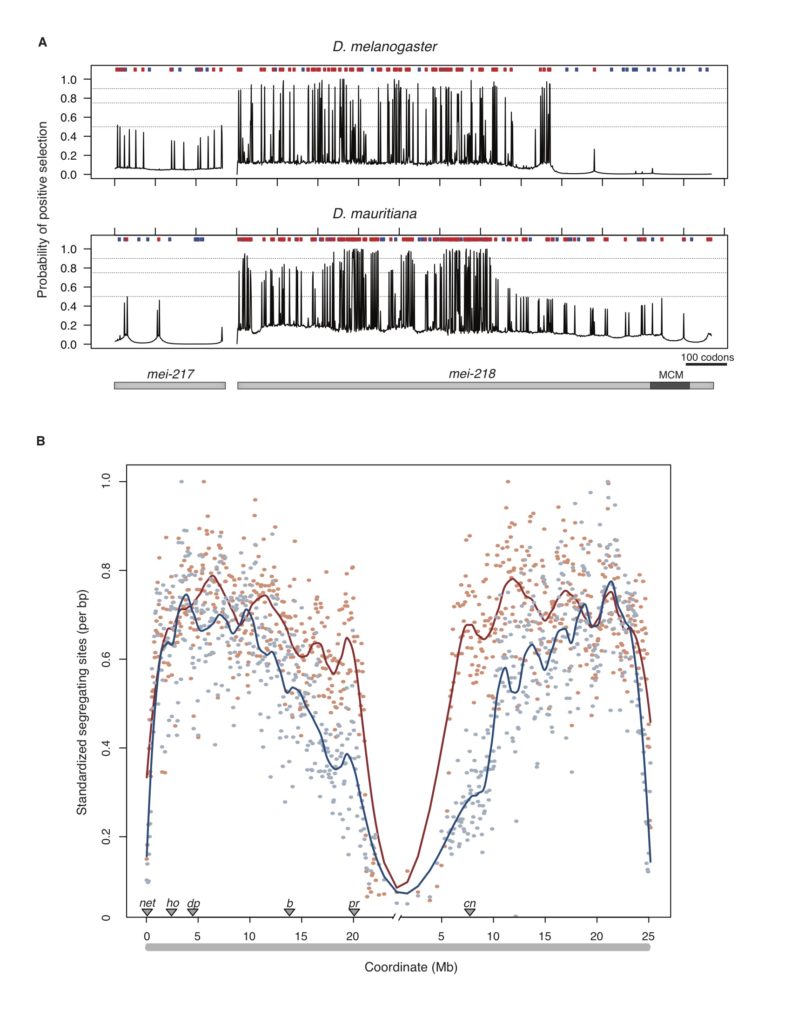
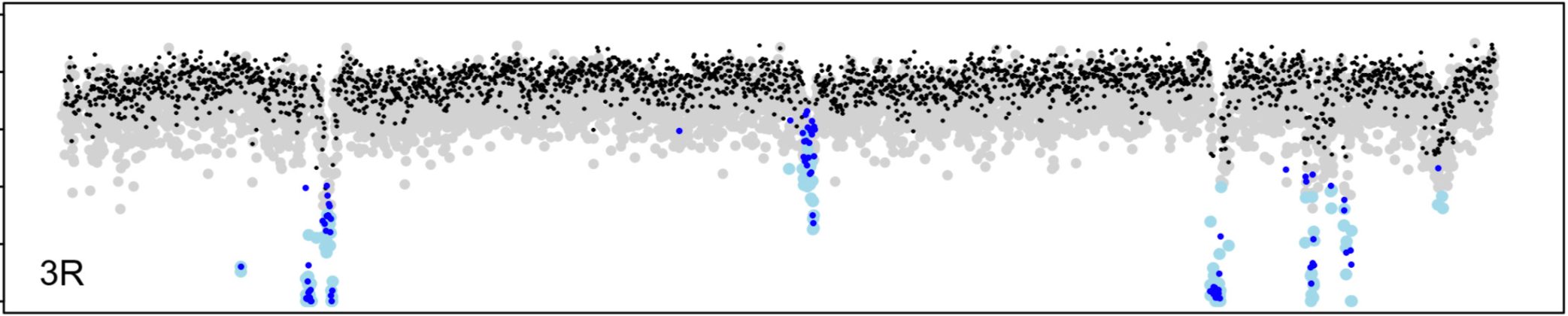
In past work, we studied how local rates of crossing over affect the efficacy of natural selection in the Drosophila genome (Betancourt and Presgraves 2002; Presgraves 2005). In our new on-going work, we are investigating the genetic control of evolved species differences in the rate of recombination. We have identified a single gene with a history of rapid evolution that can account for the entire ~1.8-fold genetic map length difference between D. melanogaster and D. mauritiana, and we’re now studying the mechanism by which such a dramatic change in recombination rate is achieved (Brand et al. 2018; Brand et al. 2019).